Mechanism research of PAK2 phosphorylating TRIM28 in the progression of prostate cancer
-
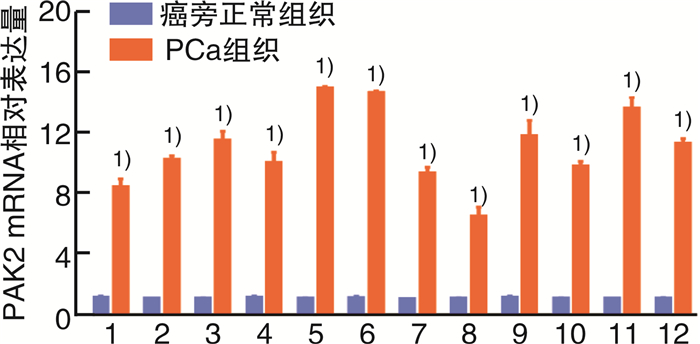
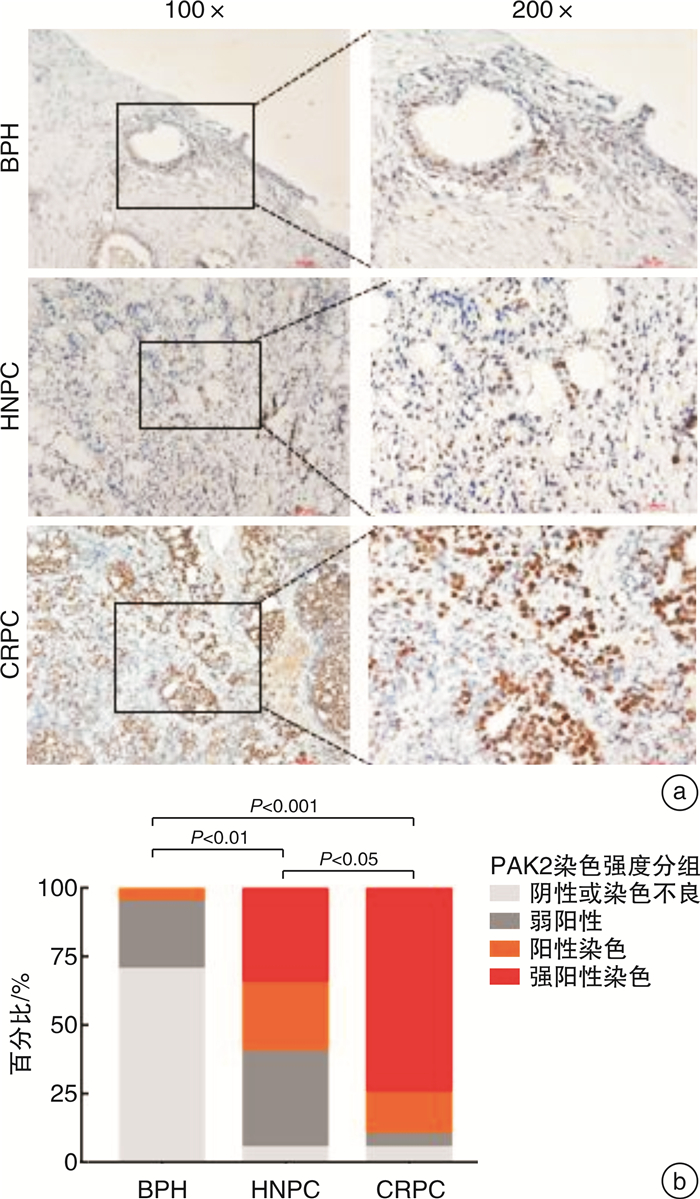
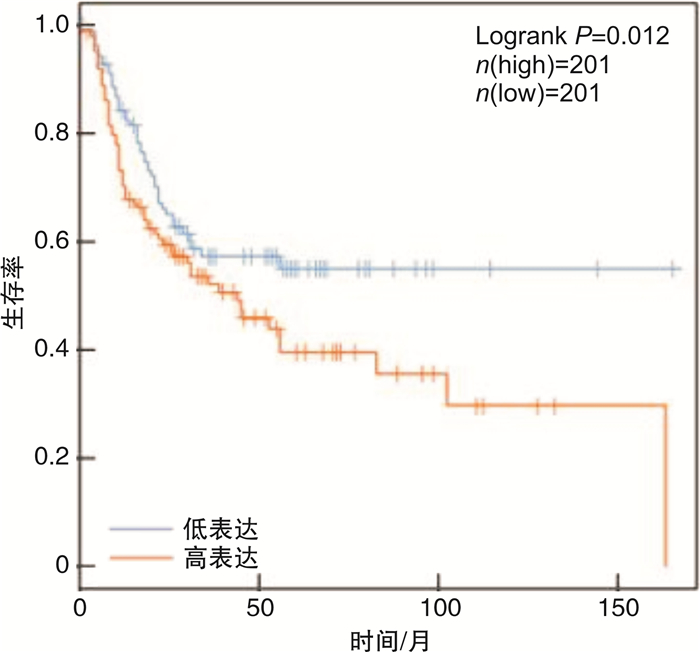
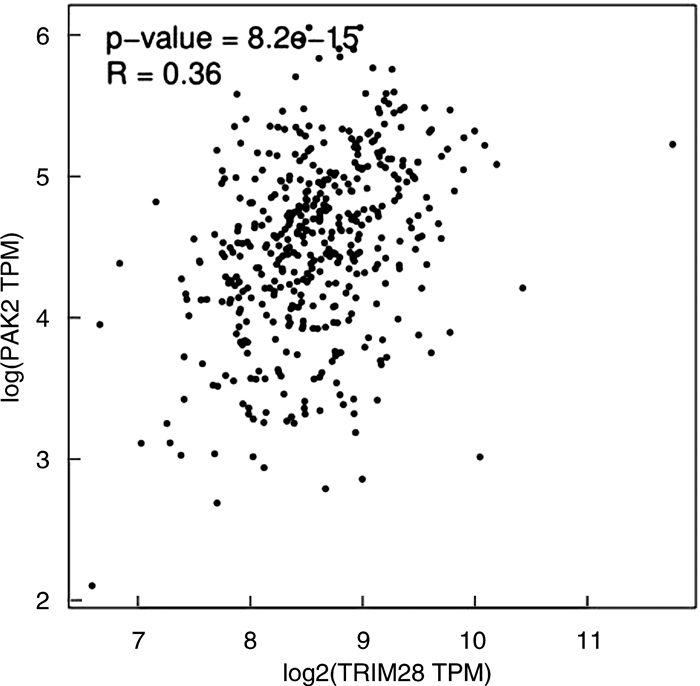
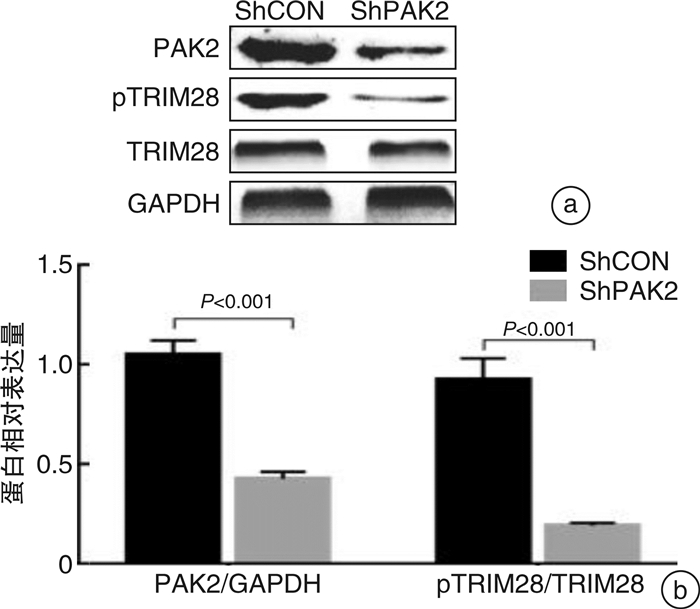
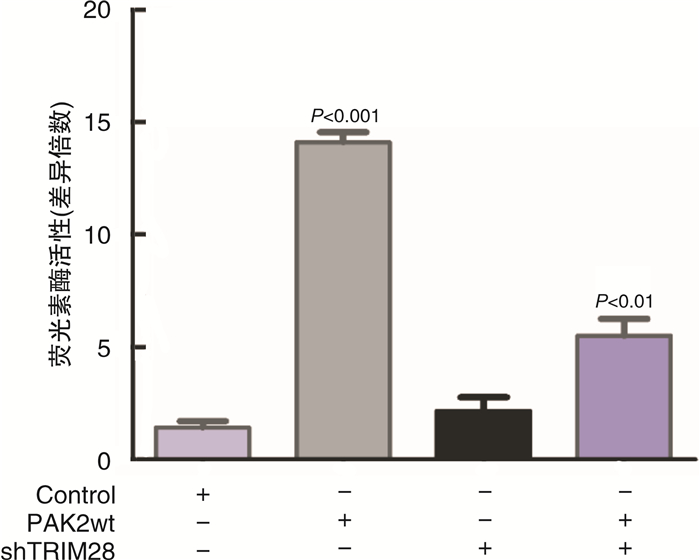
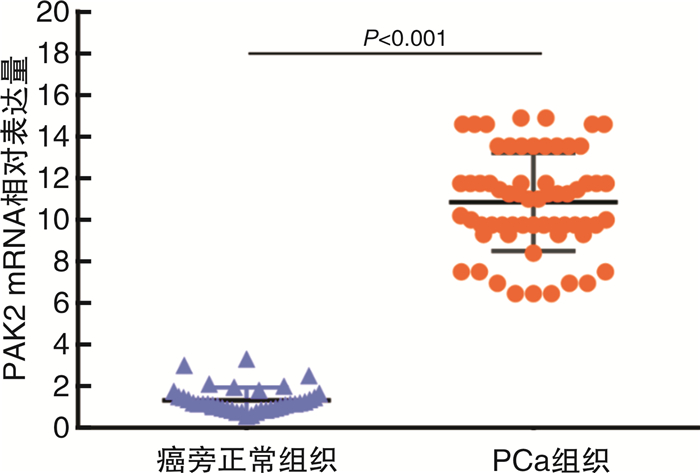
摘要: 目的 研究P21蛋白活化酶2(p21-activated protein kinase 2,PAK2)在前列腺癌(prostate cancer,PCa)中的表达水平,并分析PAK2与PCa患者临床资料及PCa进展的相关性,进一步研究及分析PAK2磷酸化激活KRAB相关蛋白1即三重基序蛋白28(tripartite motif-containing 28,TRIM28)促进PCa进展的相关性,阐明PAK2在去势抵抗性前列腺癌(castration-resistant prostate cancer,CRPC)进展中的作用关系,探讨PAK2磷酸化激活TRIM28通路在PCa进展中扮演的角色,为治疗CRPC提供方法和思路。方法 使用ONCOMINE数据库预测PAK2在PCa组织和癌旁正常组织中的表达差异。分别采用Western Blot和RT-qPCR检测PAK2在收集的临样本(癌旁正常组织和PCa组织)中的表达差异。采用免疫组织化学(immunohistochemistry,IHC)检测PAK2在前列腺增生(BPH)组织、激素性前列腺癌(androgen dependent prostate cancer,HNPC)组织和CRPC组织中的表达差异,通过癌症基因组图谱(the cancer genome atlas,TCGA)数据库预测PAK2的不同表达水平对患者疾病无进展的影响差异。使用医学统计学方法收集、整理、分析统计140例PCa患者的临床病理特征与PAK2的关系。利用GEPIA数据库预测TRIM28与PAK2在PCa中的相关性。采用Western Blot方法检测PAK2表达水平的不同对TRIM28的磷酸化水平的影响。结果 在数据库ONCOMINE中显示,PCa组织中PAK2的表达水平高于其在癌旁正常组织中的表达水平,与我们收集的临床组织中的检测结果一致。PAK2在PCa组织中的RNA表达水平高于癌旁正常组织的表达水平。同样PAK2在PCa组织中的蛋白表达水平高于癌旁组织的表达水平。IHC结果显示PAK2在BPH组织、HNPC组织和CRPC组织中的表达水平逐渐升高。TCGA数据库显示,PCa患者中PAK2低表达水平较高表达水平有一个较长的疾病无进展生存期。患者临床资料统计分析得出PAK2的表达水平与患者的PSA水平、Gleason评分、临床分期、病理分级、转移、总生存期及疾病的生化复发情况密切相关,患者的PSA水平越高,Gleason评分越高,临床分期及病理分期越高,PAK2的表达水平越高,患者越容易发生转移及生化复发,患者的生存期越短,反之亦然。GEPIA数据库显示,PAK2与TRIM28呈正向相关性。荧光素酶报告基因实验显示,只转染PAK2过表达组的SOX2启动子区活性最高。而转染PAK2过表达及敲低TRIM28组的SOX2启动子区活性低于只转染PAK2过表达组,但是明显高于对照组。结论 PAK2在PCa中表达水平显著高于癌旁正常组织,在CRPC中表达水平最高,并且PAK2的表达水平与患者的PSA水平、Gleason评分、临床分期、病理分级、转移、总生存期及疾病的生化复发情况密切相关,与患者年龄无关。PAK2表达水平可以影响SOX2的表达水平,说明PAK2可能调节肿瘤细胞的干性来影响疾病进展,PAK2可以磷酸化激活TRIM28,活化的TRIM28可以增强SOX2启动子区的活性,使肿瘤细胞的细胞学行为更为活跃。
-
关键词:
- 去势抵抗性前列腺癌 /
- 磷21蛋白活化激酶2 /
- 三元基序家族蛋白28 /
- 磷酸化 /
- 性别决定基因相关高活性群组蛋白2
Abstract: Objective In order to study the detailed expression level of PAK2 in prostate cancer, we analyzed the correlation between PAK2 and clinical data of prostate cancer patients and the progression of prostate cancer. To further study and analyze the correlation of PAK2 phosphorylation activating TRIM28 to promote the progression of prostate cancer. To elucidate the role of PAK2 in the progression of castration-resistant prostate cancer(CRPC), and explore that PAK2 phosphorylation activation of TRIM28 pathway plays a very important role in the progression of prostate cancer, providing us with new methods and ideas for the treatment of CRPC.Methods We used the ONCOMINE database to predict the difference in PAK2 expression between prostate cancer and normal prostate tissue. Western Blot and RT-qPCR were used to detect the difference of PAK2 expression in the collected clinical samples(paracancer tissue and prostate cancer tissue). The expression difference of PAK2 in benign prostatic hyperplasia(BPH) tissues, androgen dependent prostate cancer(HNPC) tissues and CRPC tissues was detected by IHC, and the influence of different expression levels of PAK2 on disease progression was predicted by TCGA database. The relationship between the clinicopathological features and PAK2 in 140 patients with prostate cancer was collected and analyzed by medical statistics. GEPIA database was used to predict the correlation between TRIM28 and PAK2 in prostate cancer. The effects of PAK2 expression levels on TRIM28 phosphorylation were detected by Western Blot.Results In the database ONCOMINE, PAK2 expression levels were shown to be higher in prostate cancer tissues than in paracancer prostate tissues, consistent with the detection results in clinical tissues collected by us. The expression level of PAK2 RNA in prostate cancer tissues was higher than that in paracancer tissues. Similarly, PAK2 protein expression level in prostate cancer tissues was higher than that in paracancer tissues. IHC results showed that PAK2 expression level increased gradually in BPH, HNPC and CRPC tissues. The TCGA database showed that patients with prostate cancer with low PAK2 expression level had a longer progression-free survival than those with high PAK2 expression level. Statistical analysis of clinical data of patients showed that the expression level of PAK2 was closely related to the PSA level, Gleason score, clinical stage, pathological grade, metastasis, overall survival and biochemical recurrence of the disease. The higher the PSA level of patients, the higher the Gleason score, and the higher the clinical stage and pathological stage, the higher the PAK2 expression level. The more likely a patient is to develop metastasis and biochemical recurrence, the shorter the patient's survival, and vice versa. GEPIA database showed that PAK2 was positively correlated with TRIM28. Luciferase reporter gene experiment showed that the activity of SOX2 promoter region was the highest in PAK2 overexpression group. The activity of SOX2 promoter in PAK2 overexpression and TRIM28 knockdown groups was lower than that in PAK2 overexpression group, but significantly higher than that in control group.Conclusion The expression level of PAK2 in prostate cancer was significantly higher than that in paracancer normal tissues, and the expression level of PAK2 was the highest in CRPC. The expression level of PAK2 was closely related to the PSA level, Gleason score, clinical stage, pathological grade, metastasis, overall survival and biochemical recurrence of the disease, but not related to the age of the patient. The expression level of PAK2 can affect the expression level of SOX2, indicating that PAK2 may regulate the dryness of tumor cells, so as to affect the progression of disease. PAK2 can phosphorylate and activate TRIM28, and activated TRIM28 can enhance the activity of SOX2 promoter region, making the cytological behavior of tumor cells more active. -

-
表 1 140例PCa患者的主要临床病理特征信息与PAK2的表达水平之间的关系
例 临床特征 例数 PAK2表达 P值 低(87例) 高(53例) 年龄/岁 0.637 < 65 76 56 20 ≥ 65 64 31 33 PSA/(ng/mL) 0.049 < 10 87 65 22 ≥10 53 22 31 Gleason评分 0.036 ≤6 101 78 23 >6 39 9 30 临床分期 0.001 ≤T2a 105 83 22 >T2a 35 4 31 病理分级 0.014 < pT3a 106 80 26 ≥pT3a 34 7 27 是否转移 0.001 否 122 84 38 是 18 3 15 总体生存 0.001 存活 131 86 45 死亡 9 1 8 是否生化复发 0.002 否 105 84 21 是 35 3 32 -
[1] Gooding AJ, Zhang B, Jahanbani FK, et al. The lncRNA BORG drives breast cancer metastasis and disease recurrence[J]. Sci Rep, 2017, 7: 12698. doi: 10.1038/s41598-017-12716-6
[2] Addison JB, Koontz C, Fugett JH, et al. KAP1 promotes proliferation and metastatic progression of breast cancer cells[J]. Cancer Res, 2015, 75(2): 344-355. doi: 10.1158/0008-5472.CAN-14-1561
[3] Cheng CT, Kuo CY, Ouyang C, et al., Metabolic Stress-Induced Phosphorylation of KAP1 Ser473 Blocks Mitochondrial Fusion in Breast Cancer Cells[J]. Cancer Res, 2016.76(17): 5006-5018. doi: 10.1158/0008-5472.CAN-15-2921
[4] El-Aarag SA, Mahmoud A, Hashem MH, et al. In silico identification of potential key regulatory factors in smoking-induced lung cancer[J]. BMC Med Genomics, 2017, 10(1): 40. doi: 10.1186/s12920-017-0284-z
[5] Fitzgerald S, Espina V, Liotta L, et al. Stromal TRIM28-associated signaling pathway modulation within the colorectal cancer microenvironment[J]. J Transl Med, 2018, 16(1): 89. doi: 10.1186/s12967-018-1465-z
[6] Fitzgerald S, Sheehan KM, O'Grady A, et al. Relationship between epithelial and stromal TRIM28 expression predicts survival in colorectal cancer patients[J]. J Gastroenterol Hepatol, 2013, 28(6): 967-974. doi: 10.1111/jgh.12157
[7] Wei CL, Cheng JL, Zhou B, et al. Tripartite motif containing 28 (TRIM28) promotes breast cancer metastasis by stabilizing TWIST1 protein[J]. Sci Rep, 2016, 6: 29822. doi: 10.1038/srep29822
[8] Grebeňová D, Holoubek A, Röselová P, et al. PAK1, PAK1Δ15, and PAK2: similarities, differences and mutual interactions[J]. Sci Rep, 2019, 9(1): 17171. doi: 10.1038/s41598-019-53665-6
[9] Hao L, Leng J, Xiao RJ, et al. Bioinformatics analysis of the prognostic value of Tripartite Motif 28 in breast cancer[J]. Oncol Lett, 2017, 13(4): 2670-2678. doi: 10.3892/ol.2017.5764
[10] Herquel B, Ouararhni K, Davidson I. The TIF1α-related TRIM cofactors couple chromatin modifications to transcriptional regulation, signaling and tumor suppression[J]. Transcription, 2011, 2(5): 231-236. doi: 10.4161/trns.2.5.17725
[11] Herquel B, Ouararhni K, Khetchoumian K, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma[J]. Proc Natl Acad Sci U S A, 2011, 108(20): 8212-8217. doi: 10.1073/pnas.1101544108
[12] Hu G, Kim J, Xu QK, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal[J]. Genes Dev, 2009, 23(7): 837-848. doi: 10.1101/gad.1769609
[13] Jov evska I, Zupanec N, Ko evar N, et al. TRIM28 and β-actin identified via nanobody-based reverse proteomics approach as possible human glioblastoma biomarkers[J]. PLoS One, 2014, 9(11): e113688. doi: 10.1371/journal.pone.0113688
[14] Li JS, Lu D, Dou H, et al. Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway[J]. Nat Commun, 2018, 9(1): 143. doi: 10.1038/s41467-017-02413-3
[15] Neo SH, Itahana Y, Alagu J, et al. TRIM28 is an E3 ligase for ARF-mediated NPM1/B23 SUMOylation that represses centrosome amplification[J]. Mol Cell Biol, 2015, 35(16): 2851-2863. doi: 10.1128/MCB.01064-14
[16] Peng Y, Zhang MM, Jiang ZZ, et al. TRIM28 activates autophagy and promotes cell proliferation in glioblastoma[J]. Onco Targets Ther, 2019, 12: 397-404. doi: 10.2147/OTT.S188101
[17] Qi ZX, Cai JJ, Chen LC, et al. TRIM28 as an independent prognostic marker plays critical roles in glioma progression[J]. J Neurooncol, 2016, 126(1): 19-26. doi: 10.1007/s11060-015-1897-8
[18] Robbez-Masson L, Tie CHC, Conde L, et al. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes[J]. Genome Res, 2018, 28(6): 836-845. doi: 10.1101/gr.228171.117
[19] Santos J, Gil J. TRIM28/KAP1 regulates senescence[J]. Immunol Lett, 2014, 162(1 Pt B): 281-289.
[20] Satoh M, Chan JYF, Ross SJ, et al. Autoantibodies to transcription intermediary factor TIF1β associated with dermatomyositis[J]. Arthritis Res Ther, 2012, 14(2): R79. doi: 10.1186/ar3802
[21] Song X, Guo CL, Zheng YT, et al. Post-transcriptional regulation of cancer/testis antigen MAGEC2 expression by TRIM28 in tumor cells[J]. BMC Cancer, 2018, 18(1): 1-10. doi: 10.1186/s12885-017-3892-2
-




 下载:
下载: