Discovery of novel biomarkers and pathways in aldosterone-producing adenomas through label-free quantitative proteomics analysis
-
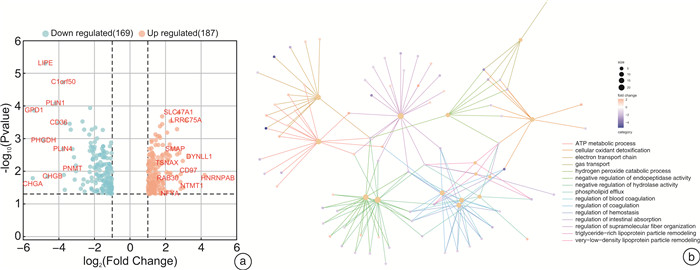
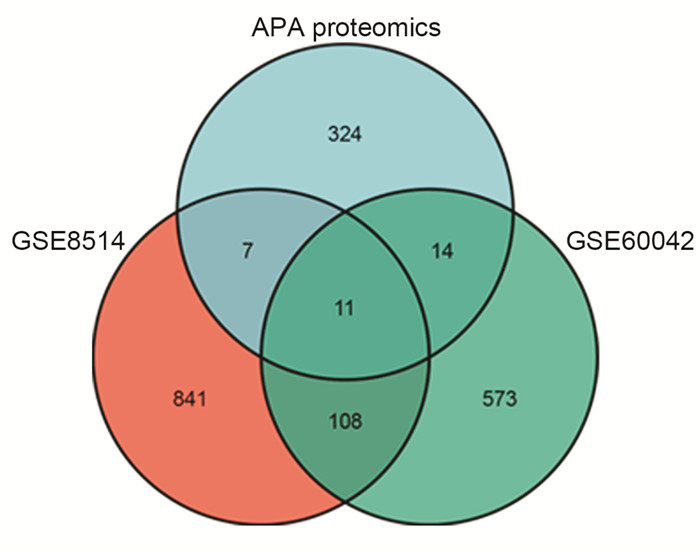
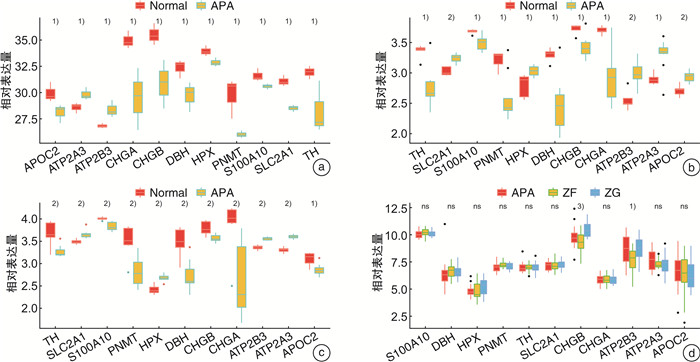
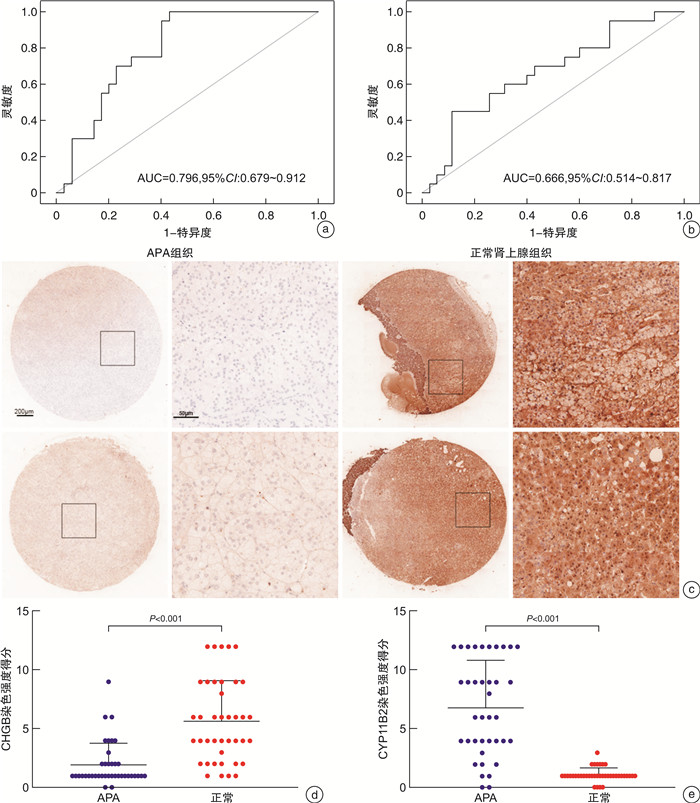
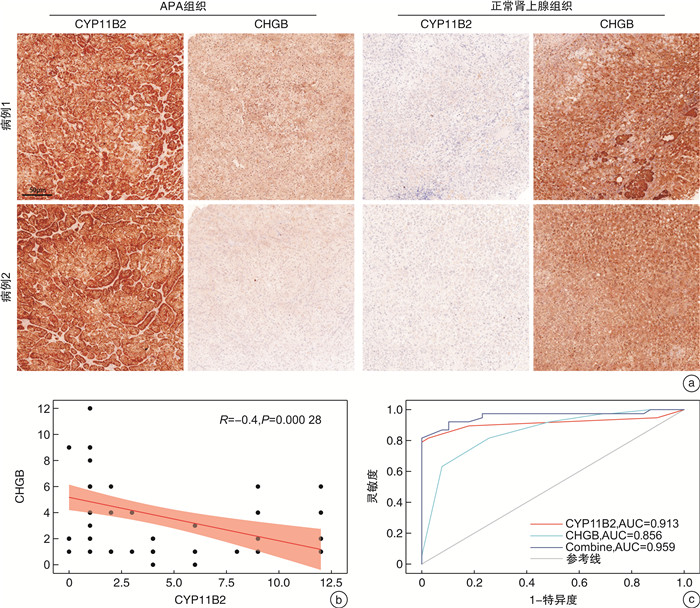
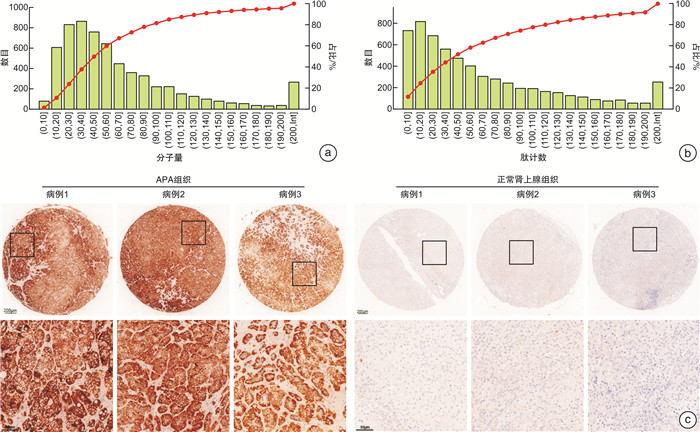
摘要: 目的 本研究旨在通过对接受肾上腺切除术的患者采集的醛固酮腺瘤(aldosterone-producing adenoma,APA)组织进行无标记定量蛋白质组学分析,筛选APA组织和正常肾上腺组织之间差异表达的蛋白。方法 利用无标记定量蛋白质组学技术对APA及正常肾上腺组织进行分析,筛选APA组织和正常肾上腺组织之间差异表达的蛋白。采用免疫组化染色对APA病人组织样本染色,验证差异表达蛋白的表达。结果 共鉴定了6 282种蛋白质。使用P < 0.05和|fold change|≥2的显著性截断值,鉴定出356种差异表达蛋白(differentially expressed proteins,DEPs)。此外,通过整合蛋白质组和转录组数据,鉴定出一个新的生物标志物嗜铬蛋白B(chromogranin B,CHGB),在APA中显著表达下调,并进一步通过免疫组化染色得到了进一步验证。受试者工作特征(receiver operating characteristic,ROC)曲线分析显示,CHGB在区分正常组织和APA方面具有较高的特异性(AUC=0.856),显示其在APA发展中的潜在重要性。重要的是,CHGB和CYP11B2的联合提高了诊断APA的预测效能。结论 我们的研究确定了参与APA发生的关键蛋白质和通路,并发现了一种新的蛋白质生物标志物来区分APA,可能成为预测APA的潜在生物标记物。这些发现为未来APA的分子诊断研究提供了理论依据。Abstract: Objective To identify differentially expressed proteins between aldosterone-producing adenoma (APA) tissue and normal adrenal tissue through label-free quantitative proteomics analysis of APA tissue collected from patients undergoing adrenal resection.Methods The APA and normal adrenal tissue were analyzed using label-free quantitative proteomics analysis to identify differentially expressed proteins. Immunohistochemical staining was used to validate the expression differences of the identified proteins between APA and normal tissue in patient samples.Results A total of 6282 proteins were identified. Using a significance cutoff of P < 0.05 and |fold change| ≥2 356 differentially expressed proteins were identified. Furthermore, through the integration of proteomics and transcriptomics data, a novel biomarker, chromogranin B(CHGB), was identified with significantly downregulated expression in APAs, which was further validated through immunohistochemical staining. ROC curve analysis showed that CHGB had high specificity in distinguishing normal tissues from APAs(AUC=0.856), indicating its potential importance in APA development. Importantly, the combination of CHGB and CYP11B2 improved the predictive efficiency for diagnosing APAs.Conclusion Our study identified key proteins and pathways involved in APA development and discovered a novel protein biomarker for distinguishing APAs, which may serve as a potential biomarker for predicting APAs. These findings provide a theoretical basis for future molecular diagnostic research in APAs.
-

-
表 1 12例APA患者的临床资料
序号 分组 性别 年龄/岁 BMI/(kg/m2) K+/(mmol/L) 醛固酮/(ng/dL) PRA/(ng/mL/h) ARR 部位 1 APA1/normal1 男 56 20.45 3.63 15.96 0.38 42 右 2 女 41 22.00 3.78 24.52 0.42 58 左 3 男 44 23.00 3.39 38.98 0.13 300 左 4 女 47 19.14 2.50 61.93 0.65 95 右 5 APA2/normal2 女 45 17.97 2.84 16.64 0.04 416 右 6 男 54 25.06 3.76 18.03 0.47 38 右 7 女 56 24.22 3.66 254.94 0.09 2 833 左 8 男 61 24.22 2.72 41.21 0.07 589 左 9 APA3/normal3 男 48 25.61 3.43 16.21 0.02 811 左 10 男 60 25.00 3.09 70.30 0.05 1 406 左 11 女 30 19.00 2.85 61.74 0.17 363 左 12 男 59 21.95 2.61 25.68 0.38 68 右 -
[1] Zavatta G, Di Dalmazi G, Pizzi C, et al. Larger ascending aorta in primary aldosteronism: a 3-year prospective evaluation of adrenalectomy vs. medical treatment[J]. Endocrine, 2019, 63(3): 470-475. doi: 10.1007/s12020-018-1801-3
[2] Hundemer GL, Curhan GC, Yozamp N, et al. Renal Outcomes in Medically and Surgically Treated Primary Aldosteronism[J]. Hypertension, 2018, 72(3): 658-666. doi: 10.1161/HYPERTENSIONAHA.118.11568
[3] Williams TA, Reincke M. Pathophysiology and histopathology of primary aldosteronism[J]. Trends Endocrinol Metab, 2022, 33(1): 36-49. doi: 10.1016/j.tem.2021.10.002
[4] Zennaro MC, Boulkroun S, Fernandes-Rosa FL. Pathogenesis and treatment of primary aldosteronism[J]. Nat Rev Endocrinol, 2020, 16(10): 578-589. doi: 10.1038/s41574-020-0382-4
[5] Xie J, Zhang C, Wang X, et al. Exploration of KCNJ5 Somatic Mutation and CYP11B1/CYP11B2 Staining in Multiple Nodules in Primary Aldosteronism[J]. Front Med(Lausanne), 2022, 9: 823065.
[6] Nanba K, Baker JE, Blinder AR, et al. Histopathology and Genetic Causes of Primary Aldosteronism in Young Adults[J]. J Clin Endocrinol Metab, 2022, 107(9): 2473-2482. doi: 10.1210/clinem/dgac408
[7] Gioco F, Seccia TM, Gomez-Sanchez EP, et al. Adrenal histopathology in primary aldosteronism: is it time for a change?[J]. Hypertension, 2015, 66(4): 724-730. doi: 10.1161/HYPERTENSIONAHA.115.05873
[8] Nam AS, Chaligne R, Landau DA. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics[J]. Nat Rev Genet, 2021, 22(1): 3-18.
[9] McGuire AL, Gabriel S, Tishkoff SA, et al. The road ahead in genetics and genomics[J]. Nat Rev Genet, 2020, 21(10): 581-596. doi: 10.1038/s41576-020-0272-6
[10] Pan Y, Lei X, Zhang Y. Association predictions of genomics, proteinomics, transcriptomics, microbiome, metabolomics, pathomics, radiomics, drug, symptoms, environment factor, and disease networks: A comprehensive approach[J]. Med Res Rev, 2022, 42(1): 441-461. doi: 10.1002/med.21847
[11] Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data[J]. Innovation(Camb), 2021, 2(3): 100141.
[12] Kanehisa M, Sato Y, Kawashima M. KEGG mapping tools for uncovering hidden features in biological data[J]. Protein Sci, 2022, 31(1): 47-53. doi: 10.1002/pro.4172
[13] Park KS, Kim SH, Oh JH, et al. Highly accurate diagnosis of papillary thyroid carcinomas based on personalized pathways coupled with machine learning[J]. Brief Bioinform, 2021, 22(4): bbaa336. doi: 10.1093/bib/bbaa336
[14] 唐俊, 彭宏伟, 代喆, 等. VHL综合征家系合并VHL基因和TP53基因突变的临床表型及基因突变分析研究[J]. 临床泌尿外科杂志, 2023, 38(11): 815-821. https://lcmw.whuhzzs.com/article/doi/10.13201/j.issn.1001-1420.2023.11.003
[15] 常智, 尚吉文, 任瑞民, 等. KCNJ5突变与单侧原发性醛固酮增多症术后临床缓解的相关性研究[J]. 临床泌尿外科杂志, 2023, 38(7): 519-524. https://lcmw.whuhzzs.com/article/doi/10.13201/j.issn.1001-1420.2023.07.008
[16] Macklin A, Khan S, Kislinger T. Recent advances in mass spectrometry based clinical proteomics: applications to cancer research[J]. Clin Proteomics, 2020, 17: 17. doi: 10.1186/s12014-020-09283-w
[17] Buzdin A, Sorokin M, Garazha A, et al. RNA sequencing for research and diagnostics in clinical oncology[J]. Semin Cancer Biol, 2020, 60: 311-323. doi: 10.1016/j.semcancer.2019.07.010
[18] Fedosova NU, Habeck M, Nissen P. Structure and Function of Na, K-ATPase-The Sodium-Potassium Pump[J]. Compr Physiol, 2021, 12(1): 2659-2679.
[19] Scholl UI. Genetics of Primary Aldosteronism[J]. Hypertension, 2022, 79(5): 887-897. doi: 10.1161/HYPERTENSIONAHA.121.16498
[20] Yoshida Y, Shibata H. Recent progress in the diagnosis and treatment of primary aldosteronism[J]. Hypertens Res, 2023, 46(7): 1738-1744. doi: 10.1038/s41440-023-01288-w
[21] Pitsava G, Faucz FR, Stratakis CA, et al. Update on the Genetics of Primary Aldosteronism and Aldosterone-Producing Adenomas[J]. Curr Cardiol Rep, 2022, 24(9): 1189-1195. doi: 10.1007/s11886-022-01735-z
[22] Yadav GP, Wang H, Ouwendijk J, et al. Chromogranin B(CHGB)is dimorphic and responsible for dominant anion channels delivered to cell surface via regulated secretion[J]. Front Mol Neurosci, 2023, 16: 1205516. doi: 10.3389/fnmol.2023.1205516
[23] Watanabe T. The Emerging Roles of Chromogranins and Derived Polypeptides in Atherosclerosis, Diabetes, and Coronary Heart Disease[J]. Int J Mol Sci, 2021, 22(11): 6118. doi: 10.3390/ijms22116118
[24] Miki M, Ito T, Hijioka M, et al. Utility of chromogranin B compared with chromogranin A as a biomarker in Japanese patients with pancreatic neuroendocrine tumors[J]. Jpn J Clin Oncol, 2017, 47(6): 520-528.
-




 下载:
下载: