Clinical study on the distribution of ESBL genotypes in drug-resistant escherichia coli in urinary tract infection
-
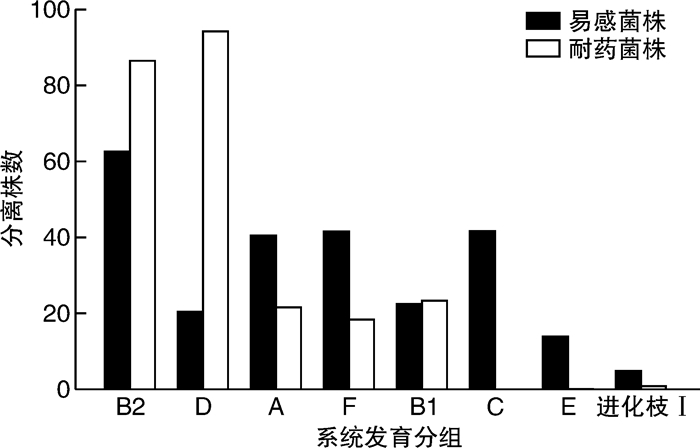
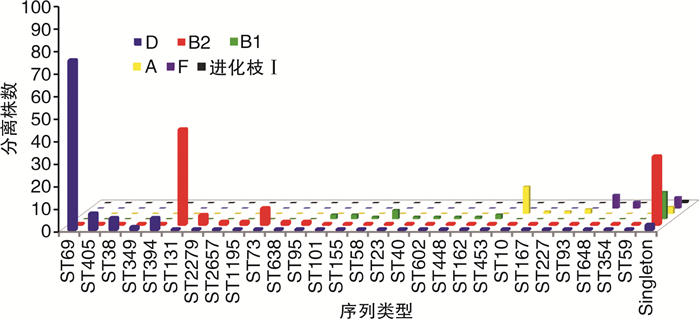
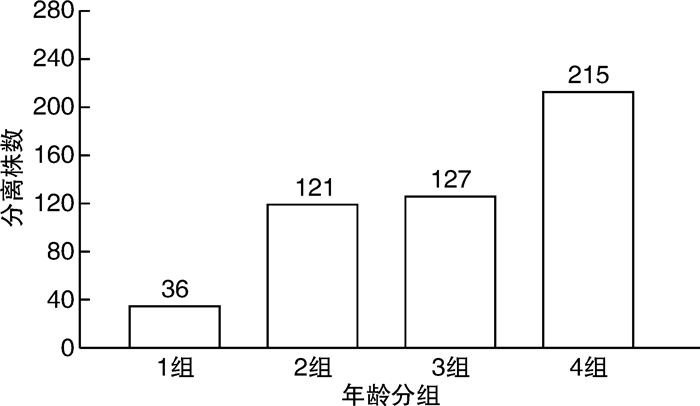
摘要: 目的 对从尿路感染(urinary tract infections,UTI)中分离出的非超广谱β-内酰胺酶(extended-spectrum β-lactamase,ESBL)和产生ESBL的大肠杆菌菌株进行基因分型,并确定尿路致病性大肠杆菌(uropathogenic Escherichia coli,UPEC)菌株谱系。方法 从南京市第一医院的临床实验室收集来自UTI诊断患者尿液的499株大肠杆菌。分离株来自2018年11月—2022年3月不同病房,包括泌尿科、肾脏科、ICU、内科、儿科的住院患者。使用氨苄西林抗性大肠杆菌分离株进行聚合酶链反应(polymerase chain reaction,PCR),以研究β-内酰胺酶基因。通过多重PCR检测对所有分离株进行多位点序列分型(multilocus sequence typing,MLST)。结果 在244株(49%)分离株中发现了对氨苄西林、环丙沙星或甲氧苄啶-磺胺甲恶唑的耐药性;128株(26%)分离株是多药耐药性(multidrug resistance,MDR),包括34株(7%)产ESBL菌株。在>59岁的受试者中分离MDR大肠杆菌和产ESBL大肠杆菌的概率较其他年龄组明显增加(P < 0.01)。MLST分析显示,耐药性分离株主要属于系统发育组D和B2的ST。其中,CC69(75株)最常见,所有分离株都包括在ST69中(75株,100%)。其次是CC131(48株),包括ST131(42株,86%)、ST2279(4株,8%)、ST2657(1株,2%)和ST1195(1株,2%)。255株易感菌株中,仅检出2株ST69和3株ST131。在34株产生ESBL的分离株中,31株(91%)含有blaCTX-M型基因;17株(55%)含blaCTX-M-15,9株(29%)含blaCTX-M-8。结论 从UTI中检测肠杆菌科细菌具有较高的ESBL发生率,并且对常用的抗生素的耐药性增加。耐药性的增加与属于ST69和ST131的UPEC分离株有关。Abstract: Objective Genotyping of non-extended-spectrum β-lactamase(ESBL) and ESBL-producing Escherichia coli strains isolated from urinary tract infections(UTIs) and determination of uropathogenic Escherichia coli(UPEC) strain lineages.Methods Four hundred and ninety-nine strains of E.coli from the urine of UTI-diagnosed patients were collected from the clinical laboratory of Nanjing First Hospital. The isolates were obtained from different wards of our hospital from November 2018 to March 2022, including inpatients in urology, nephrology, ICU, internal medicine, pediatrics. Polymerase chain reaction(PCR) was performed using ampicillin-resistant E.coli isolates to study the beta-lactamase gene. All isolates were subjected to multilocus sequence typing(MLST) by multiplex PCR assay.Results Drug resistance to ampicillin, ciprofloxacin, or trimethoprim-sulfamethoxazole was found in 244(49%) isolates; 128(26%) isolates were multidrug resistant(MDR), including 34(7%) ESBL-producing strains. The probability of isolation of MDR E.coli and ESBL-producing E.coli was significantly higher in subjects >59 years old than those in other age groups(P < 0.01). MLST analysis showed that the drug-resistant isolates mainly belonged to STs of phylogeny D and B2. Among them, CC69(n=75) was the most common, and all isolates were included in ST69(n=75, 100%). CC131(n=48), including ST131(n=42, 86%), ST2279(n=4, 8%), ST2657(n=1, 2%), and ST1195(n=1, 2%) followed. Among the 255 susceptible strains, only 2 ST69 strains and 3 ST131 strains were detected. Of the 34 ESBL-producing isolates, 31(91%) contained the blaCTX-M type gene; 17(55%) contained blaCTX-M-15, and 9(29%) contained blaCTX-M-8.Conclusion Enterobacteriaceae detected from UTI has a high incidence of ESBL and increased resistance to commonly used antibiotics. Increased resistance is associated with UPEC isolates belonging to ST69 and ST131.
-

-
表 1 499株大肠杆菌中的抗生素耐药性按患者年龄分类
株(%) 抗生素 按年龄划分的耐药菌株 P值 < 18岁(36株) 18~39岁(121株) 40~59岁(127株) >59岁(215株) 氨苄西林 16(44.44) 41(33.88) 47(37.01) 103(47.91) 0.007 头孢噻吩 5(13.89) 29(23.97) 43(33.86) 93(43.26) < 0.001 萘啶酸 4(11.11) 21(16.54) 30(23.62) 92(42.79) < 0.001 甲氧苄啶-磺胺甲恶唑 8(22.22) 22(18.18) 33(25.98) 85(39.53) < 0.001 环丙沙星 2(5.56) 10(8.26) 16(12.60) 70(32.56) < 0.001 诺氟沙星 3(8.33) 10(8.26) 18(14.17) 68(31.63) < 0.001 头孢呋辛 4(11.11) 13(10.74) 18(14.17) 54(25.12) 0.002 阿莫西林-克拉维酸 4(11.11) 7(5.79) 11(8.66) 37(17.21) 0.006 头孢曲松 1(2.78) 3(2.48) 7(5.51) 34(15.81) < 0.001 头孢吡肟 1(2.78) 4(3.31) 7(5.51) 32(14.88) < 0.001 庆大霉素 1(2.78) 3(2.48) 4(3.15) 26(12.09) 0.002 呋喃妥因 1(2.78) 10(8.26) 3(2.36) 13(6.05) 0.300 阿米卡星 1(2.78) 0(0) 2(1.57) 4(1.86) 0.250 MDR 5(13.89) 17(14.05) 27(21.26) 79(36.74) < 0.001 ESBL 0(0) 1(0.83) 3(2.36) 30(13.95) 0.001 表 2 34株产生ESBL和173株不产生ESBL的大肠杆菌分离株中的β-内酰胺酶编码基因和序列类型
CC/ST(株) β-内酰胺酶基因 株(%) 产生ESBL的大肠杆菌分离株 131/131(9) blaCTX-M-15 5(56) blaCTX-M-15+blaCMY-2 1(11) blaCTX-M-27+blaCMY-2 1(11) blaCTX-M-2 1(11) blaCTX-M-8 1(11) 648/648(5) blaCTX-M-15+blaTEM-1 3(60) blaCTX-M-15 2(40) 405/405(3) blaCTX-M-15+blaTEM-1 3(100) 155/155(2) blaCTX-M-15+blaCMY-2 1(50) blaTEM-1+blaSHV-2 1(50) 38/38(2) blaCTX-M-14(1);blaCTX-M-17(1) 1(50);1(50) 73/73(1) blaCTX-M-1 1 Singleton(12) blaCTX-M-15+blaTEM-1(2);blaCTX-M-8+blaTEM-1(3) blaCTX-M-8(5);blaSHV-2(2) 2(17);3(25) 5(42);2(17) 不产生ESBL的大肠杆菌分离株 69/69(62) blaTEM 53(85) 131/131(27) blaTEM 24(89) 131/2279(4);131/2657(1) blaTEM 5(100) 131/1195(1) blaTEM+blaCMY-2 1 10/10(10) blaTEM 9(90) 10/227(1) blaTEM 1 73/73(6) blaTEM 6(100) 73/638(1) blaSHV-1 1 648/648(1) blaCMY-2 1 394/394(5) blaTEM 5(100) 38/38(3) blaTEM 3(100) 101/101(2) blaTEM 2(100) 405/405(4);168/93(2);23/23(2);59/59(2);86/453(1);155/155(1);349/564(1) blaTEM 13(100) Singleton(36) blaTEM type 32(89) -
[1] 孔娜娜, 张培燕, 杨德平, 等. 上海周浦地区2016年至2020年尿路感染者病原体分布情况及耐药性检测[J]. 诊断学理论与实践, 2021, 20(1): 93-97.
[2] 黄晶晶, 叶颖子, 俞蕙, 等. 上海地区单中心0至14岁儿童尿路感染的病原菌分布及耐药分析[J]. 中华传染病杂志, 2022, 40(2): 71-78.
[3] Nabadda S, Kakooza F, Kiggundu R, et al. Implementation of the World Health Organization global antimicrobial resistance surveillance system in Uganda, 2015-2020: mixed-methods study using national surveillance data[J]. JMIR Public Health Surveill, 2021, 7(10): e29954. doi: 10.2196/29954
[4] 何卓琳, 唐敏嘉, 蒲万霞. 大肠杆菌分子分型方法研究进展[J]. 中国草食动物科学, 2021, 41(5): 57-60.
[5] Manges AR, Geum HM, Guo A, et al. Global extraintestinal pathogenic Escherichia coli(ExPEC)lineages[J]. Clin Microbiol Rev, 2019, 32(3): e00135-18.
[6] Matinfar S, Ahmadi M, Sisakht AM, et al. Phylogenetic and antibiotics resistance in extended-spectrum B-lactamase(ESBL)uropathogenic Escherichia coli: an update review[J]. Gene Rep, 2021, 23: 101168. doi: 10.1016/j.genrep.2021.101168
[7] 张韶华, 葛藤, 金萍, 等. 南京市哨点医院96株沙门菌血清分型及药敏试验结果分析[J]. 医学动物防制, 2022, 38(3): 221-225.
[8] Tayh G, Al Laham N, Fhoula I, et al. Frequency and Antibiotics Resistance of Extended-Spectrum Beta-Lactamase(ESBLs)Producing Escherichia coli and Klebsiella pneumoniae Isolated from Patients in Gaza Strip, Palestine[J]. J Med Microbiol, 2021, 9(3): 133-141.
[9] Kang CI, Kim J, Park DW, et al. Clinical practice guidelines for the antibiotic treatment of community-acquired urinary tract infections[J]. Infect Chemother, 2018, 50(1): 67-100.
[10] 陈中举, 田磊, 杨为民, 等. 2016—2018年泌尿外科患者尿路感染病原菌分布及耐药性分析[J]. 临床泌尿外科杂志, 2020, 35(2): 103-107, 111. https://lcmw.whuhzzs.com/article/doi/10.13201/j.issn.1001-1420.2020.02.004
[11] 张钻兵, 李美珍, 邱琳. 某院2018年—2019年泌尿系结石患者尿路感染病原菌的分布及其耐药性分析[J]. 抗感染药学, 2021, 18(7): 1018-1021.
[12] van Driel AA, Notermans DW, Meima A, et al. Antibiotic resistance of Escherichia coli isolated from uncomplicated UTI in general practice patients over a 10-year period[J]. Eur J Clin Microbiol Infect Dis, 2019, 38(11): 2151-2158.
[13] Flament-Simon SC, Nicolas-Chanoine MH, García V, et al. Clonal structure, virulence factor-encoding genes and antibiotic resistance of Escherichia coli, causing urinary tract infections and other extraintestinal infections in humans in Spain and France during 2016[J]. Antibiotics, 2020, 9(4): 161.
[14] Nascimento JAS, Santos FF, Santos-Neto JF, et al. Molecular epidemiology and presence of hybrid pathogenic Escherichia coli among isolates from community-acquired urinary tract infection[J]. Microorganisms, 2022, 10(2): 302.
[15] Yamaji R, Rubin, Thys E, et al. Persistent pandemic lineages of uropathogenic Escherichia coli in a college community from 1999 to 2017[J]. J Clin Microbiol, 2018, 56(4): e01834-17.
-




 下载:
下载: