Identification of key genes in the tumor microenvironment of renal clear cell carcinoma and establishment of a nomogram
-
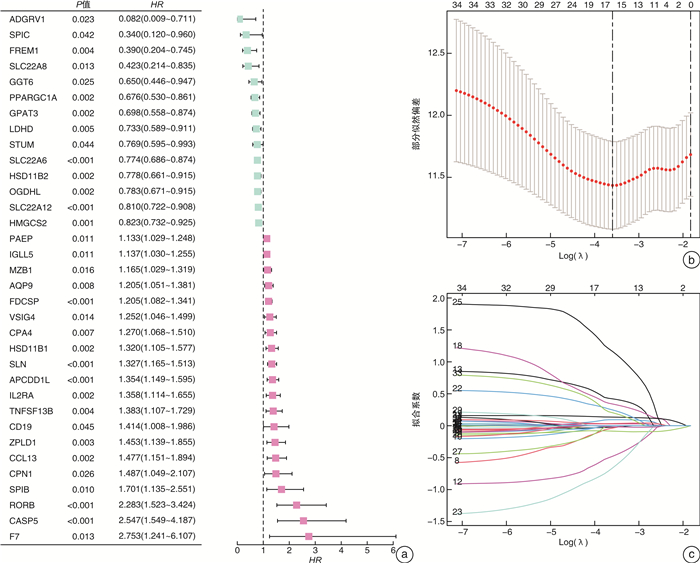
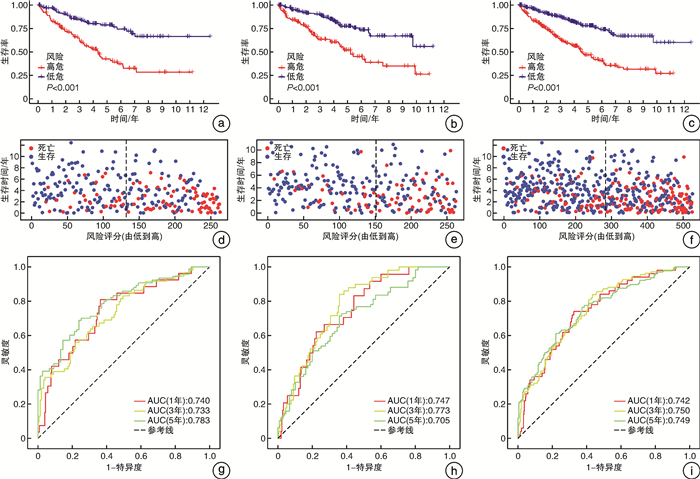
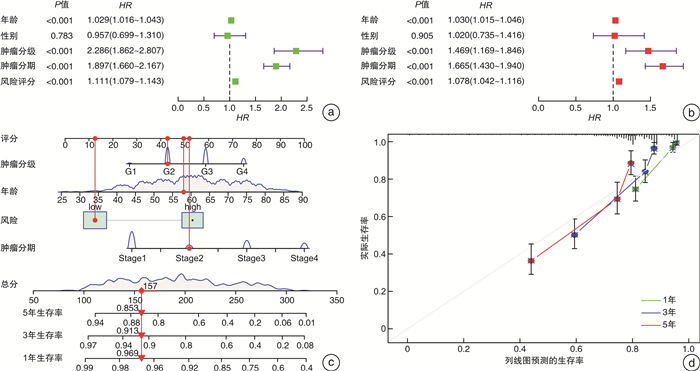
摘要: 目的 筛选肾透明细胞癌(clear cell renal cell carcinoma,ccRCC)微环境中的关键基因,并建立预后预测模型。方法 从TCGA数据库中下载ccRCC RNA-seq FPKM数据及临床数据。基于ESTIMATE算法对肿瘤微环境中免疫细胞、基质细胞进行评分,以免疫细胞,基质细胞评分中位值为界把样品各自分组,组间进行差异表达基因(DEGs)分析,并用wilcoxon检验进行验证,对得到的DEGs取交集获得肿瘤微环境中的关键基因。对获得的关键基因进行功能富集。把样品按照1:1的比例分为训练组和验证组。利用Cox-LASSO法从训练组中的关键基因中筛选出建模基因。多因素Cox回归建立风险评估模型,将风险评估模型与临床指标进行单因素和多因素Cox回归筛选患者预后的独立影响因素。联合各项患者预后独立相关的因素绘制列线图。结果 免疫细胞评分组共获得DEGs 658个,而利用基质细胞评分共获得DEGs 411个,取交集后共得到95个关键基因。随机分组后,训练组与验证组性别、年龄、肿瘤分期及分级的构成比较均差异无统计学意义。训练组中34个基因与患者预后相关,进而通过LASSO回归获得15个特征基因。经多因素Cox分析得到由9个基因(HMGCS2、FREM1、CASP5、SLN、SPIC、SPIB、RORB、CPN1、F7)构成的最优模型。生存分析表明训练组与验证组高低风险组间生存率的差异有统计学意义(P < 0.001),且各组对于1、3、5年生存率的评估有着较高的灵敏度与特异度。经单因素和多因素Cox回归分析发现患者年龄、肿瘤分期、分级、风险评分为患者预后的独立影响因素(均P < 0.001)。联合患者临床因素如年龄、肿瘤分级和肿瘤分期及风险评分构建了预后评估的列线图模型。结论 由9个基因构成的风险评分模型可对ccRCC风险做出准确判断;风险评分、年龄、肿瘤分期、肿瘤分级为ccRCC独立预后影响因素,由其构成的列线图模型可精准预测患者生存率。Abstract: Objective To construct a risk prediction model for patients with clear cell renal cell carcinoma(ccRCC) and create a nomogram for survival prediction.Methods RNA-seq FPKM data and clinical data of ccRCC were downloaded from TCGA database. Based on the ESTIMATE algorithm, immune cells and stromal cells in the tumor microenvironment were scored; The samples were grouped by the median value of immune cells and stromal cells, and the differentially expressed genes(DEGs) were analyzed between groups and validated by wilcoxon test. Functional enrichment was performed on the obtained key genes. The samples were divided into a training group and a validation group in a 1∶1 ratio. Modeling genes were screened from the key genes in the training group using the Cox-LASSO method. Multi-factor Cox regression was used to establish risk assessment models, and univariate and multi-factor Cox regressions were performed to screen independent influences on patient prognosis with clinical indicators. The factors independently associated with each patient prognosis were combined to draw a nomogram.Results A total of 658 DEGs were obtained according to the immune cell scoring group, while a total of 411 DEGs were obtained using stromal cell scoring, and a total of 95 key genes were obtained after taking the intersection. After randomization, there were no significant differences in the composition of gender, age, tumor stage or grade between the training and validation groups. Of these, 34 genes were associated with patient prognosis, and then 15 signature genes were obtained by LASSO regression. The optimal model consisting of 9 genes(HMGCS2, FREM1, CASP5, SLN, SPIC, SPIB, RORB, CPN1, F7) was obtained by multifactorial Cox analysis. Survival analysis showed significant differences in survival between the high and low risk groups in the training and validation groups(P < 0.001), and the groups had high sensitivity and specificity for the assessment of survival at 1, 3 and 5 years. Univariate and multifactorial Cox regression analysis revealed patients' age, tumor stage, grade and risk score as independent influences on patients' prognosis(all P < 0.001). Nomogram models were constructed by combining patient clinical factors such as age, tumor grade and tumor stage and risk score.Conclusion The risk score model composed by 9 genes can accurately evaluate the risk of ccRCC; the risk score, age, tumor stage, and tumor grade are independent prognostic factors for ccRCC, and the nomogram composed by them can accurately predict the survival rate of the patients.
-
Key words:
- clear cell renal cell carcinoma /
- tumor microenvironment /
- nomogram
-

-
表 1 训练组及验证组数据分布
例(%) 变量 总计(526例) 训练组(263例) 验证组(263例) P值 年龄/岁 0.462 ≤65 347(65.97) 178(67.68) 169(64.26) > 65 179(34.03) 85(32.32) 94(35.74) 性别 1.000 女 183(34.79) 92(34.98) 91(34.60) 男 343(65.21) 171(65.02) 172(65.40) 肿瘤分级 0.476 G1 13(2.47) 7(2.66) 6(2.28) G2 226(42.97) 113(42.97) 113(42.97) G3 205(38.97) 95(36.12) 110(41.83) G4 74(14.07) 42(15.97) 32(12.17) 未知 8(1.52) 6(2.28) 2(0.76) 肿瘤分期 0.336 Ⅰ 261(49.62) 141(53.61) 120(45.63) Ⅱ 57(10.84) 25(9.51) 32(12.17) Ⅲ 123(23.38) 58(22.05) 65(24.71) Ⅳ 82(15.59) 38(14.45) 44(16.73) 未知 3(0.57) 1(0.38) 2(0.76) -
[1] Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma[J]. Nat Rev Dis Primers, 2017, 3: 17009. doi: 10.1038/nrdp.2017.9
[2] Liu D, Shu GF, Jin FY, et al. ROS-responsive chitosan-SS31 prodrug for AKI therapy via rapid distribution in the kidney and long-term retention in the renal tubule[J]. Sci Adv, 2020, 6(41): eabb7422. doi: 10.1126/sciadv.abb7422
[3] Jonasch E, Walker CL, Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality[J]. Nat Rev Nephrol, 2021, 17(4): 245-261. doi: 10.1038/s41581-020-00359-2
[4] Bussard KM, Mutkus L, Stumpf K, et al. Tumor-associated stromal cells as key contributors to the tumor microenvironment[J]. Breast Cancer Res, 2016, 18(1): 84. doi: 10.1186/s13058-016-0740-2
[5] Tian CX, Huang Y, Clauser KR, et al. Suppression of pancreatic ductal adenocarcinoma growth and metastasis by fibrillar collagens produced selectively by tumor cells[J]. Nat Commun, 2021, 12(1): 2328. doi: 10.1038/s41467-021-22490-9
[6] Li XY, Wenes M, Romero P, et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy[J]. Nat Rev Clin Oncol, 2019, 16(7): 425-441. doi: 10.1038/s41571-019-0203-7
[7] Turajlic S, Swanton C, Boshoff C. Kidney cancer: the next decade[J]. J Exp Med, 2018, 215(10): 2477-2479. doi: 10.1084/jem.20181617
[8] Arjumand W, Sultana S. Role of VHL gene mutation in human renal cell carcinoma[J]. Tumour Biol, 2012, 33(1): 9-16. doi: 10.1007/s13277-011-0257-3
[9] Chen FJ, Zhang YQ, Şenbabaoǧlu Y, et al. Multilevel genomics-based taxonomy of renal cell carcinoma[J]. Cell Rep, 2016, 14(10): 2476-2489. doi: 10.1016/j.celrep.2016.02.024
[10] Fukumura D, Kloepper J, Amoozgar Z, et al. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges[J]. Nat Rev Clin Oncol, 2018, 15(5): 325-340. doi: 10.1038/nrclinonc.2018.29
[11] Massari F, Rizzo A, Mollica V, et al. Immune-based combinations for the treatment of metastatic renal cell carcinoma: a meta-analysis of randomised clinical trials[J]. Eur J Cancer, 2021, 154: 120-127. doi: 10.1016/j.ejca.2021.06.015
[12] Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy[J]. Cell, 2017, 168(4): 707-723. doi: 10.1016/j.cell.2017.01.017
[13] Carlsson R, Persson C, Leanderson T. SPI-C, a PU-box binding ETS protein expressed temporarily during B-cell development and in macrophages, contains an acidic transactivation domain located to the N-terminus[J]. Mol Immunol, 2003, 39(16): 1035-1043. doi: 10.1016/S0161-5890(03)00032-4
[14] Alam Z, Devalaraja S, Li MH, et al. Counter regulation of spic by NF-κB and STAT signaling controls inflammation and iron metabolism in macrophages[J]. Cell Rep, 2020, 31(13): 107825. doi: 10.1016/j.celrep.2020.107825
[15] Du W, Xu X, Niu Q, et al. Spi-B-mediated silencing of claudin-2 promotes early dissemination of lung cancer cells from primary tumors[J]. Cancer Res, 2017, 77(18): 4809-4822. doi: 10.1158/0008-5472.CAN-17-0020
[16] Martinon F, Tschopp J. [J]. Cell Death Differ, 2007, 14(1): 10-22.
[17] Soung YH, Jeong EG, Ahn CH, et al. Mutational analysis of caspase 1, 4, and 5 genes in common human cancers[J]. Hum Pathol, 2008, 39(6): 895-900. doi: 10.1016/j.humpath.2007.10.015
[18] Zhang Y, Chen XW, Fu QH, et al. Comprehensive analysis of pyroptosis regulators and tumor immune microenvironment in clear cell renal cell carcinoma[J]. Cancer Cell Int, 2021, 21(1): 667. doi: 10.1186/s12935-021-02384-y
[19] Han PP, Wang YF, Luo WQ, et al. Epigenetic inactivation of hydroxymethylglutaryl CoA synthase reduces ketogenesis and facilitates tumor cell motility in clear cell renal carcinoma[J]. Pathol Res Pract, 2021, 227: 153622. doi: 10.1016/j.prp.2021.153622
[20] Kashem MA, Li HZ, Liu LR, et al. The potential role of FREM1 and its isoform TILRR in HIV-1 acquisition through mediating inflammation[J]. Int J Mol Sci, 2021, 22(15): 7825. doi: 10.3390/ijms22157825
[21] Wang GG, Wang Z, Lu HQ, et al. Comprehensive analysis of FRAS1/FREM family as potential biomarkers and therapeutic targets in renal clear cell carcinoma[J]. Front Pharmacol, 2022, 13: 972934. doi: 10.3389/fphar.2022.972934
-




 下载:
下载: