Development and validation of cuproptosis-related genes expression-based signature to predict overall survival of clear cell renal cell carcinoma
-
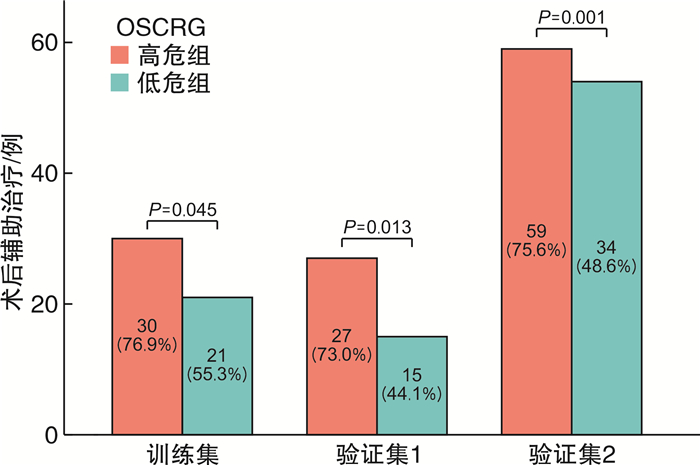
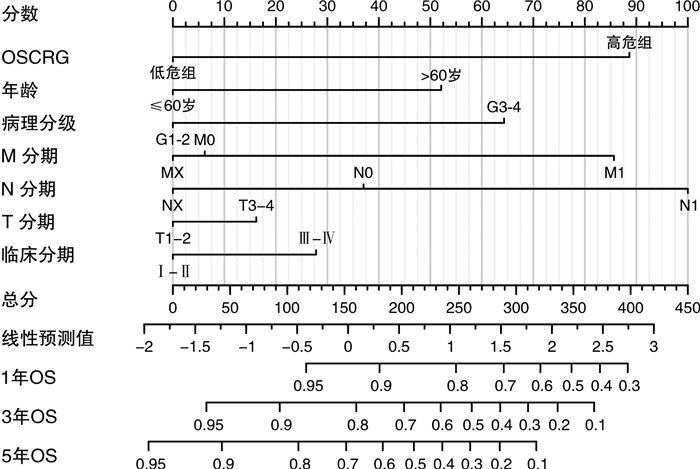
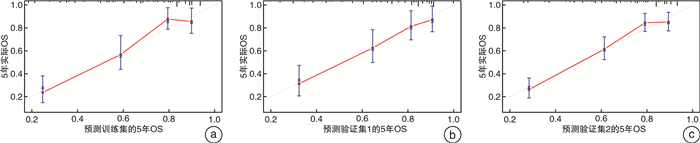
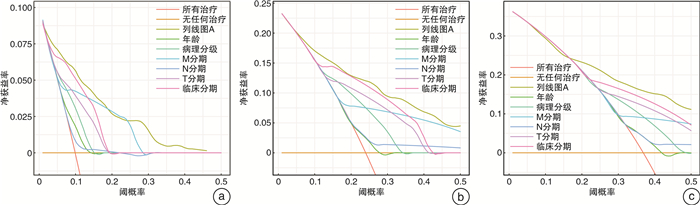
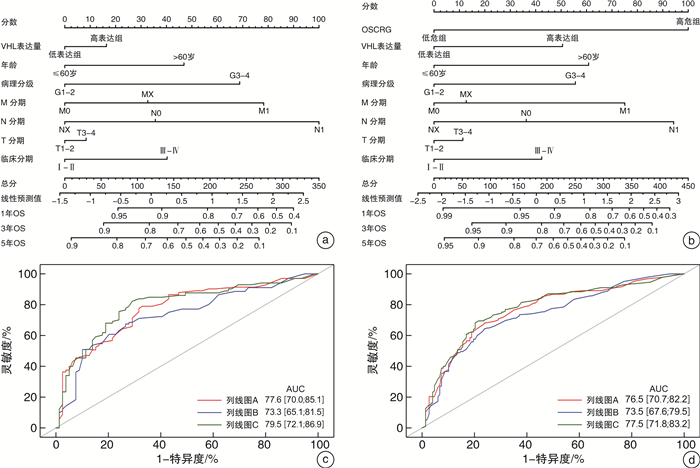
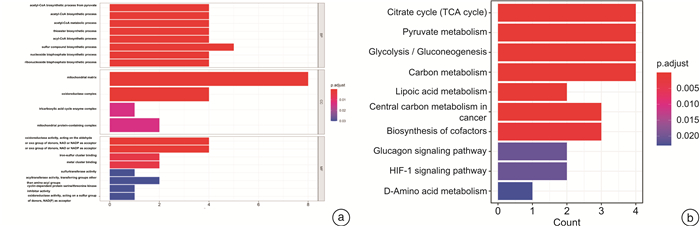
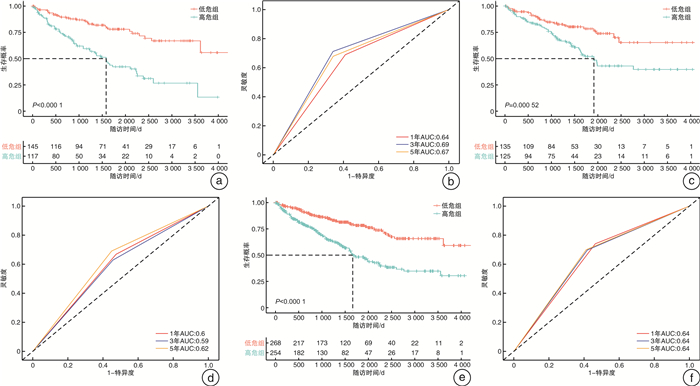
摘要: 目的 研究铜死亡相关基因(cuproposis-related genes,CRGs)在肾透明细胞癌(clear cell renal cell carcinoma,ccRCC)中的表达情况,构建预后模型并探讨其临床意义。方法 从TCGA数据库中获得522例ccRCC患者的转录组数据和临床病理数据。将患者随机分为训练集(262例)和验证集1(260例),并将总体患者作为验证集2(522例)。在训练集中采用差异性分析确定肿瘤组织和癌旁组织差异性表达的CRGs,使用LASSO-Cox回归分析构建ccRCC的CRGs预后模型(OSCRG)。根据OSCRG将患者分为低危组和高危组,使用Kaplan-Meier生存分析研究OSCRG与总体生存期(overall survival,OS)的关系。使用单因素和多因素Cox回归分析构建包含OSCRG和临床病理参数的列线图预测OS,并在验证集中验证该列线图的准确性。最后使用KEGG和GO基因富集分析探索差异性表达的CRGs的生物学功能。结果 在训练集中有9个差异性表达的CRGs,并且均与OS相关。LASSO-Cox回归分析确定了3个CRGs并构建OSCRG。使用OSCRG将患者分为高危组和低危组,生存分析显示在训练集和验证集中低危组OS均高于高危组,并且OSCRG是OS的独立预后因素。使用OSCRG和临床病理参数构建的列线图对OS具有良好的预测作用,且加入VHL表达量后其预测能力增强。此外,在训练集和验证集中,高危组术后接受辅助治疗的患者多于低危组。最后,GO和KEGG分析结果表明,差异性表达的CRGs与代谢相关。结论 3种CRGs可能是ccRCC的预后分子标志物,有望为患者的个体化治疗提供新的参考依据。Abstract: Objective To clarify an expression pattern of cuproposis-related genes(CRGs) in individual overall survival(OS) risk evaluation among patients with clear cell renal cell carcinoma(ccRCC).Methods In this study, 522 patients with ccRCC from the TCGA database were included. Differentially expressed genes(DEGs) were detected in a training cohort(n=262) to establish a gene classifier by Cox regression model. The accuracy of prognostic prediction of this gene classifier was validated in two validation cohorts(n=260 and n=522, respectively). Additionally, we conducted Kyoto Encyclopedia of Genes and Genomes(KEGG) and Gene Ontology(GO) analyses to assess the biological functions of the differentially expressed CRGs.Results In the training cohort, a total of nine differentially expressed CRGs were associated with OS. An overall survival CRGs signature(OSCRG) consisting of three genes was developed to separate patients into high-risk and low-risk groups in the training cohort. In the training cohort, individuals with low-risk scores had longer OS(P < 0.0001) than those with high-risk scores. We validated the prognostic accuracy of OSCRG in the two validation cohorts. Moreover, our results showed that patients with additional therapy in the high-risk group were more than those in the low-risk group. We developed a nomogram on the basis of the OSCRG and other variables that predicted an individual's OS risk, which was enhanced by adding VHL expression. The outcomes of GO and KEGG analyses showed that differentially expressed CRGs were metabolic-related.Conclusion Three CRGs may be molecular markers to predict the prognosis of ccRCC, and useful to guide individualized therapeutic decisions.
-
Key words:
- renal cell carcinoma /
- clear cell renal cell carcinoma /
- cuproptosis /
- signature
-

-
表 1 训练集和验证集患者的临床病理信息
例,X±S,M(IQR) 项目 训练集
(262例)验证集1
(260例)验证集2
(522)年龄/岁 59.9±11.9 61.1±12.1 60.5±12.0 性别 男 182 159 341 女 80 101 181 随访时间/d 1 173 (1 458) 1 264 (1 318) 1 200 (1 371) 生存状态 存活 171 180 351 死亡 91 80 171 术后辅助治疗 是 51 42 93 否 26 29 55 NA 185 189 374 病理分级 Gx 3 2 5 G1 6 7 13 G2 112 111 223 G3 103 101 204 G4 37 37 74 NA 1 2 3 M分期 Mx 17 11 28 M0 207 207 414 M1 37 41 78 NA 1 1 2 N分期 Nx 140 128 268 N0 116 122 238 N1 6 10 16 T分期 T1 132 133 265 T2 40 28 68 T3 86 92 178 T4 4 7 11 临床分期 Ⅰ期 130 129 259 Ⅱ期 31 25 56 Ⅲ期 61 61 122 Ⅳ期 40 42 82 NA - 3 3 OSFRG 低危组 142 149 291 高危组 120 111 231 表 2 多因素Cox回归分析
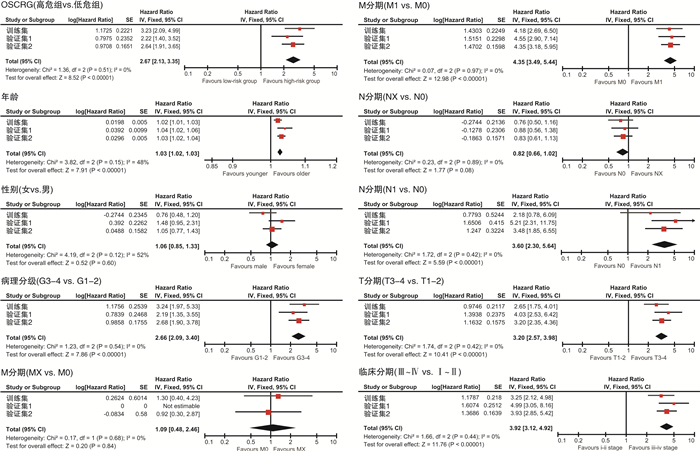
项目 训练集 验证集1 验证集2 HR(95%CI) P值 HR(95%CI) P值 HR(95%CI) P值 OSCRG 高危组 Ref 低危组 0.39(0.25~0.60) <0.000 1 0.51(0.31~0.83) 0.006 7 0.44(0.32~0.62) <0.000 1 年龄 1.03(1.01~1.05) 0.005 0 1.03(1.01~1.06) 0.002 6 1.03(1.02~1.05) <0.000 1 性别 女 Ref 男 1.41(0.87~2.29) 0.159 3 0.60(0.36~1.00) 0.048 0 0.98(0.70~1.36) 0.888 3 病理分级 G1~2 Ref G3~4 2.03(1.19~3.45) 0.008 9 1.20(0.70~2.07) 0.502 3 1.60(1.10~2.32) 0.014 0 T分期 T1~2 Ref T3~4 1.12(0.50~2.53) 0.780 7 0.78(0.29~2.11) 0.624 2 0.91(0.50~1.68) 0.765 3 N分期 N0 Ref N1 2.04(0.65~6.38) 0.218 5 1.06(0.41~2.76) 0.902 6 1.51(0.76~3.00) 0.237 1 Nx 0.64(0.41~0.99) 0.045 9 0.91(0.55~1.50) 0.713 6 0.78(0.56~1.07) 0.126 5 M分期 M0 Ref M1 2.62(1.47~4.67) 0.001 0 2.72(1.53~4.81) 0.000 6 2.46(1.67~3.63) <0.000 1 Mx 0.88(0.27~2.87) 0.835 2 0(0~int) 0.995 6 0.81(0.26~2.58) 0.722 5 临床分期 Ⅰ~Ⅱ Ref Ⅲ~Ⅳ 1.53(0.59~4.01) 0.383 9 3.10(1.01~9.50) 0.048 0 2.17(1.07~4.37) 0.030 9 -
[1] Singh D. Current updates and future perspectives on the management of renal cell carcinoma[J]. Life Sci, 2021, 264: 118632. doi: 10.1016/j.lfs.2020.118632
[2] Obradovic A, Chowdhury N, Haake SM, et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages[J]. Cell, 2021, 184(11): 2988-3005. e16. doi: 10.1016/j.cell.2021.04.038
[3] Deng T, He ZH, Duan XL, et al. STAM prolongs clear cell renal cell carcinoma patients' survival via inhibiting cell growth and invasion[J]. Front Oncol, 2021, 11: 611081. doi: 10.3389/fonc.2021.611081
[4] Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins[J]. Science, 2022, 375(6586): 1254-1261. doi: 10.1126/science.abf0529
[5] Shao W, Yang ZC, Fu Y, et al. The pyroptosis-related signature predicts prognosis and indicates immune microenvironment infiltration in gastric cancer[J]. Front Cell Dev Biol, 2021, 9: 676485. doi: 10.3389/fcell.2021.676485
[6] Zhuang W, Chen JB, Li YN, et al. Valuation of lymph node dissection in localized high-risk renal cell cancer using X-tile software[J]. Int Urol Nephrol, 2020, 52(2): 253-262. doi: 10.1007/s11255-019-02307-x
[7] Su W, He BH, Zhang YD, et al. C-index regression for recurrent event data[J]. Contemp Clin Trials, 2022, 118: 106787. doi: 10.1016/j.cct.2022.106787
[8] Van Calster B, Wynants L, Verbeek JFM, et al. Reporting and interpreting decision curve analysis: a guide for investigators[J]. Eur Urol, 2018, 74(6): 796-804. doi: 10.1016/j.eururo.2018.08.038
[9] Tang XR, Li YQ, Liang SB, et al. Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: a retrospective, multicentre, cohort study[J]. Lancet Oncol, 2018, 19(3): 382-393. doi: 10.1016/S1470-2045(18)30080-9
[10] Thakur T, Batra I, Luthra M, et al. Gene expression-assisted cancer prediction techniques[J]. J Healthc Eng, 2021, 2021: 4242646.
[11] Pleasance E, Bohm A, Williamson LM, et al. Whole-genome and transcriptome analysis enhances precision cancer treatment options[J]. Ann Oncol, 2022, 33(9): 939-949. doi: 10.1016/j.annonc.2022.05.522
[12] Yu XC, Cao S, Zhou Y, et al. Co-expression based cancer staging and application[J]. Sci Rep, 2020, 10(1): 10624. doi: 10.1038/s41598-020-67476-7
[13] Li YM, Jin JS, Bai F. Cancer biology deciphered by single-cell transcriptomic sequencing[J]. Protein Cell, 2022, 13(3): 167-179. doi: 10.1007/s13238-021-00868-1
[14] Xu WH, Xu Y, Wang J, et al. Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment[J]. Aging, 2019, 11(17): 6999-7020. doi: 10.18632/aging.102233
[15] Chang KL, Yuan C, Liu XG. Ferroptosis-related gene signature accurately predicts survival outcomes in patients with clear-cell renal cell carcinoma[J]. Front Oncol, 2021, 11: 649347. doi: 10.3389/fonc.2021.649347
[16] Zhang ZD, Lin EY, Zhuang HK, et al. Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma[J]. Cancer Cell Int, 2020, 20: 27. doi: 10.1186/s12935-020-1113-6
[17] Wang S, Chen SM, Ying YF, et al. Comprehensive analysis of ferroptosis regulators with regard to PD-L1 and immune infiltration in clear cell renal cell carcinoma[J]. Front Cell Dev Biol, 2021, 9: 676142. doi: 10.3389/fcell.2021.676142
[18] Yin XM, Wang ZY, Wang JF, et al. Development of a novel gene signature to predict prognosis and response to PD-1 blockade in clear cell renal cell carcinoma[J]. Oncoimmunology, 2021, 10(1): 1933332. doi: 10.1080/2162402X.2021.1933332
[19] Rappold PM, Vuong L, Leibold J, et al. A targetable myeloid inflammatory state governs disease recurrence in clear-cell renal cell carcinoma[J]. Cancer Discov, 2022, 12(10): 2308-2329. doi: 10.1158/2159-8290.CD-21-0925
[20] Gill DM, Hahn AW, Hale P, et al. Overview of current and future first-line systemic therapy for metastatic clear cell renal cell carcinoma[J]. Curr Treat Options Oncol, 2018, 19(1): 6. doi: 10.1007/s11864-018-0517-1
[21] Lalani AK A, McGregor BA, Albiges L, et al. Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: current paradigms, use of immunotherapy, and future directions[J]. Eur Urol, 2019, 75(1): 100-110. doi: 10.1016/j.eururo.2018.10.010
[22] Chen YL, Jiang SJ, Lu ZY, et al. Development and verification of a nomogram for prediction of recurrence-free survival in clear cell renal cell carcinoma[J]. J Cell Mol Med, 2020, 24(2): 1245-1255. doi: 10.1111/jcmm.14748
[23] Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European association of urology guidelines on renal cell carcinoma: the 2019 update[J]. Eur Urol, 2019, 75(5): 799-810. doi: 10.1016/j.eururo.2019.02.011
[24] Tsvetkov P, Detappe A, Cai K, et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress[J]. Nat Chem Biol, 2019, 15(7): 681-689. doi: 10.1038/s41589-019-0291-9
[25] Schulz V, Basu S, Freibert SA, et al. Functional spectrum and specificity of mitochondrial ferredoxins FDX1 and FDX2[J]. Nat Chem Biol, 2023, 19(2): 206-217. doi: 10.1038/s41589-022-01159-4
[26] Shi YB, Ghosh M, Kovtunovych G, et al. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis[J]. Biochim Biophys Acta, 2012, 1823(2): 484-492. doi: 10.1016/j.bbamcr.2011.11.002
[27] Zhang ZY, Ma YR, Guo XL, et al. FDX1 can impact the prognosis and mediate the metabolism of lung adenocarcinoma[J]. Front Pharmacol, 2021, 12: 749134. doi: 10.3389/fphar.2021.749134
[28] Barresi V, Trovato-Salinaro A, Spampinato G, et al. Transcriptome analysis of copper homeostasis genes reveals coordinated upregulation of SLC31A1, SCO1, and COX11 in colorectal cancer[J]. FEBS Open Bio, 2016, 6(8): 794-806. doi: 10.1002/2211-5463.12060
[29] Ji L, Zhao GN, Zhang P, et al. Knockout of MTF1 inhibits the epithelial to mesenchymal transition in ovarian cancer cells[J]. J Cancer, 2018, 9(24): 4578-4585. doi: 10.7150/jca.28040
[30] El-Mokadem I, Lim A, Kidd T, et al. Microsatellite alteration and immunohistochemical expression profile of chromosome 9p21 in patients with sporadic renal cell carcinoma following surgical resection[J]. BMC Cancer, 2016, 16: 546. doi: 10.1186/s12885-016-2514-8
[31] Pomerantz J, Schreiber-Agus N, Liégeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53[J]. Cell, 1998, 92(6): 713-723. doi: 10.1016/S0092-8674(00)81400-2
-




 下载:
下载: